NMR is the only technique capable of providing unique insights into protein dynamics and conformational changes as well as structural information at the atomic level. Detailed structures of small proteins can be elucidated using uniform 13C and 15N labeling. For larger systems (MW < 40 kDa), deuteration helps reducing spin diffusion and decreases 13C and 15N relaxation rates. However, the use of perdeuteration results in the suppression of long-range Nuclear Overhauser Effects (NOEs) arising from aliphatic and aromatic protons that are crucial for structure determination.
Stereo Array Isotope Labeling (SAIL) is an approach developed by Prof. Kainosho in 2006 that opens the way to NMR structure elucidation of large proteins (up to 50 kDa) at the atomic level. The original technology exploits stereo-selective 13C and 2H labeling of amino acids to limit spin diffusion, reduce signal overlapping and allow stereospecific assignment of prochiral groups.
Stereo-selective 13C and 2H labeling of SAIL amino acids is based on the following strategy:
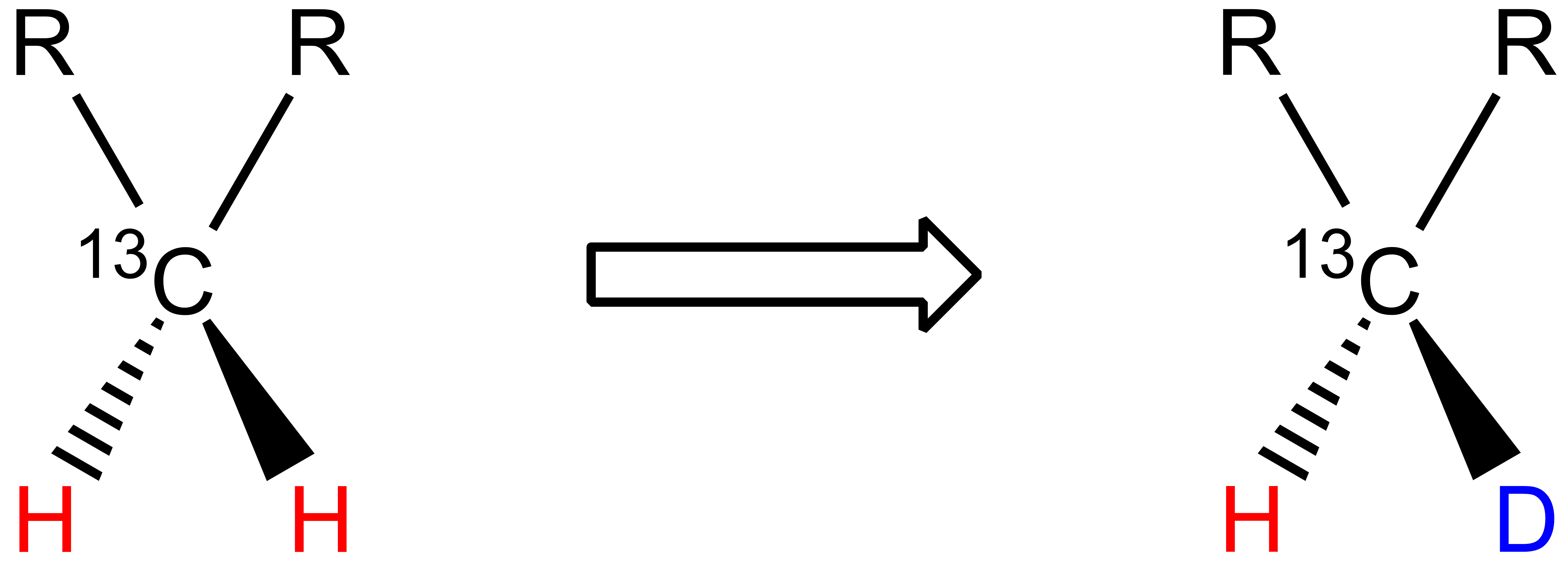
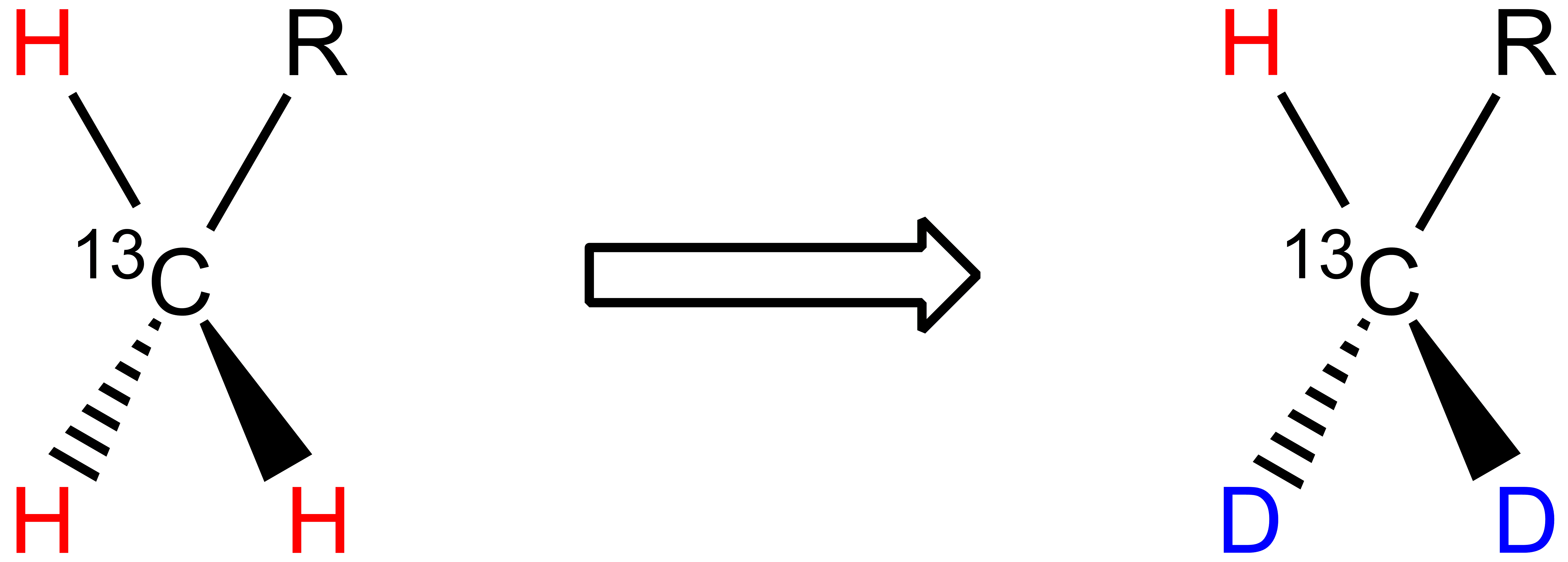
One 1H atom is replaced by one 2H atom in methylene groups:
Two 1H atoms are replaced by two 2H atoms in methyl groups:
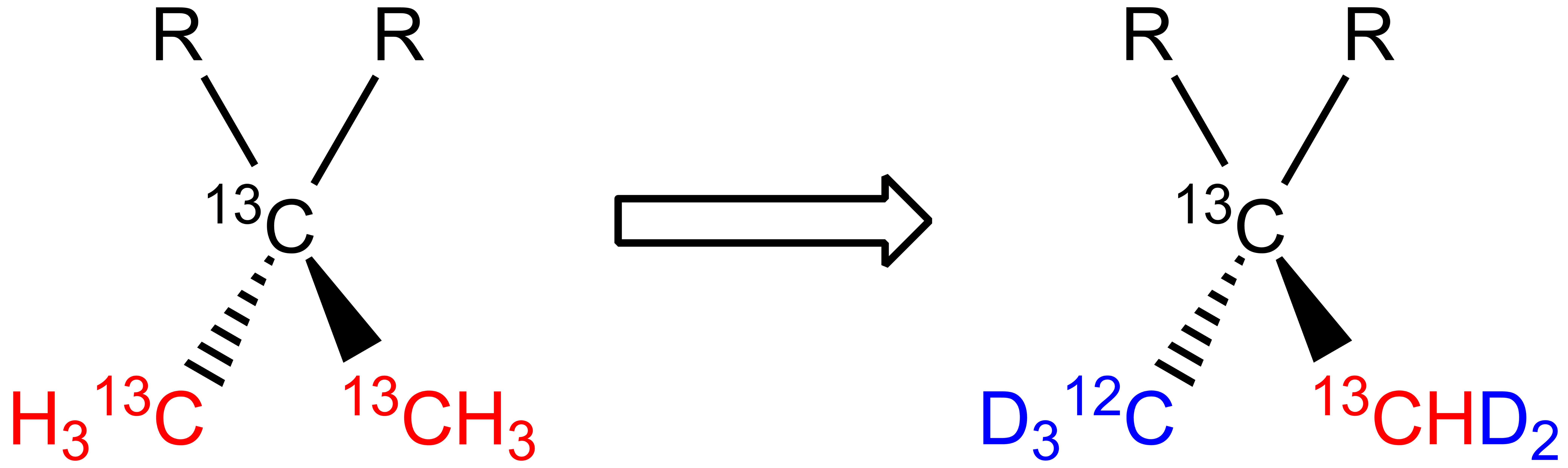
One specific methyl is labeled as 12CD3, thus kept “NMR silent” in Leucine and Valine prochiral methyl groups:
Aromatic rings are labeled with alternative 12CD and 13CH moieties.
The SAIL strategy is well-adapted to NMR applications for several reasons:
- The preservation of through-bond connectivity and the stereo-specific labeling of protons enables stereospecific backbone and side-chain NMR assignments;
- Reduction of spin diffusion enables accurate determination of distances through Nuclear Overhauser Effects (NOEs);
- Removal of 2JHH couplings and significant reduction of 3JHH couplings;
- The reduction of the number of protons in the system leads to significant improvements of NMR spectra quality;
- The reduction overlapping signals on NMR spectra enables automation of the assignments and structure calculations.
The original SAIL approach can be employed for structural and dynamics investigations on proteins produced in bacteria or insect cell and is particularly adapted to cell-free ex¬pression systems. For larger biological systems (up to 1000 kDa), TROSY-SAIL methods combining the use of SAIL amino acids and TROSY- NMR techniques have been developped and help providing unique information on protein conformation.

Cortec Isotope is pleased to offer a wide selection of SAIL amino acids available for direct use. Custom-made SAIL amino acids mixtures containing selectively labeled methyl and aromatic amino acids are available on request.
Cortec Isotope proposes to help you label your protein. For inquiry, please contact our labeled protein expression platform.